2. Huang Example
Let’s go over the Huang Bayesian Belief Network (BBN) [HD99].
2.1. Model
The Huang BBN is stored in a JSON file. For completeness, we show the content of the JSON file. Notice that the top-level JSON keys are d (directed acyclic graph) and p (parameters). Under the d the top-level keys are nodes and edges. The top-level keys under p are the names of the variables.
[1]:
import json
with open("../../_support/bbn/huang-bbn.json", "r") as fp:
data = json.load(fp)
data
[1]:
{'d': {'nodes': ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H'],
'edges': [['A', 'B'],
['A', 'C'],
['B', 'D'],
['C', 'E'],
['C', 'G'],
['D', 'F'],
['E', 'F'],
['E', 'H'],
['G', 'H']]},
'p': {'A': {'columns': ['A', '__p__'], 'data': [['on', 0.5], ['off', 0.5]]},
'B': {'columns': ['A', 'B', '__p__'],
'data': [['on', 'on', 0.5],
['on', 'off', 0.5],
['off', 'on', 0.4],
['off', 'off', 0.6]]},
'C': {'columns': ['A', 'C', '__p__'],
'data': [['on', 'on', 0.7],
['on', 'off', 0.3],
['off', 'on', 0.2],
['off', 'off', 0.8]]},
'D': {'columns': ['B', 'D', '__p__'],
'data': [['on', 'on', 0.9],
['on', 'off', 0.1],
['off', 'on', 0.5],
['off', 'off', 0.5]]},
'E': {'columns': ['C', 'E', '__p__'],
'data': [['on', 'on', 0.3],
['on', 'off', 0.7],
['off', 'on', 0.6],
['off', 'off', 0.4]]},
'F': {'columns': ['D', 'E', 'F', '__p__'],
'data': [['on', 'on', 'on', 0.01],
['on', 'on', 'off', 0.99],
['on', 'off', 'on', 0.01],
['on', 'off', 'off', 0.99],
['off', 'on', 'on', 0.01],
['off', 'on', 'off', 0.99],
['off', 'off', 'on', 0.99],
['off', 'off', 'off', 0.01]]},
'G': {'columns': ['C', 'G', '__p__'],
'data': [['on', 'on', 0.8],
['on', 'off', 0.2],
['off', 'on', 0.1],
['off', 'off', 0.9]]},
'H': {'columns': ['E', 'G', 'H', '__p__'],
'data': [['on', 'on', 'on', 0.05],
['on', 'on', 'off', 0.95],
['on', 'off', 'on', 0.95],
['on', 'off', 'off', 0.05],
['off', 'on', 'on', 0.95],
['off', 'on', 'off', 0.05],
['off', 'off', 'on', 0.95],
['off', 'off', 'off', 0.05]]}}}
From this BBN-specified JSON file, we will create the capable reasoning model.
[2]:
from pybbn.factory import create_reasoning_model
model = create_reasoning_model(data["d"], data["p"])
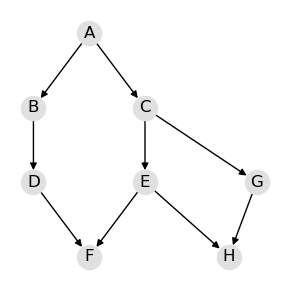
2.1.1. Graph (DAG)
We can visualize the directed acyclic graph (DAG) of the Huang BBN.
[3]:
from help.viz import *
draw_dag(model)
2.1.2. Parameters
Here are the parameters of the Huang BBN.
[4]:
model.node_potentials["A"]
[4]:
| A | __p__ | |
|---|---|---|
| 0 | on | 0.5 |
| 1 | off | 0.5 |
[5]:
model.node_potentials["B"]
[5]:
| A | B | __p__ | |
|---|---|---|---|
| 0 | on | on | 0.5 |
| 1 | on | off | 0.5 |
| 2 | off | on | 0.4 |
| 3 | off | off | 0.6 |
[6]:
model.node_potentials["C"]
[6]:
| A | C | __p__ | |
|---|---|---|---|
| 0 | on | on | 0.7 |
| 1 | on | off | 0.3 |
| 2 | off | on | 0.2 |
| 3 | off | off | 0.8 |
[7]:
model.node_potentials["D"]
[7]:
| B | D | __p__ | |
|---|---|---|---|
| 0 | on | on | 0.9 |
| 1 | on | off | 0.1 |
| 2 | off | on | 0.5 |
| 3 | off | off | 0.5 |
[8]:
model.node_potentials["E"]
[8]:
| C | E | __p__ | |
|---|---|---|---|
| 0 | on | on | 0.3 |
| 1 | on | off | 0.7 |
| 2 | off | on | 0.6 |
| 3 | off | off | 0.4 |
[9]:
model.node_potentials["F"]
[9]:
| D | E | F | __p__ | |
|---|---|---|---|---|
| 0 | on | on | on | 0.01 |
| 1 | on | on | off | 0.99 |
| 2 | on | off | on | 0.01 |
| 3 | on | off | off | 0.99 |
| 4 | off | on | on | 0.01 |
| 5 | off | on | off | 0.99 |
| 6 | off | off | on | 0.99 |
| 7 | off | off | off | 0.01 |
[10]:
model.node_potentials["G"]
[10]:
| C | G | __p__ | |
|---|---|---|---|
| 0 | on | on | 0.8 |
| 1 | on | off | 0.2 |
| 2 | off | on | 0.1 |
| 3 | off | off | 0.9 |
[11]:
model.node_potentials["H"]
[11]:
| E | G | H | __p__ | |
|---|---|---|---|---|
| 0 | on | on | on | 0.05 |
| 1 | on | on | off | 0.95 |
| 2 | on | off | on | 0.95 |
| 3 | on | off | off | 0.05 |
| 4 | off | on | on | 0.95 |
| 5 | off | on | off | 0.05 |
| 6 | off | off | on | 0.95 |
| 7 | off | off | off | 0.05 |
2.2. Graphs
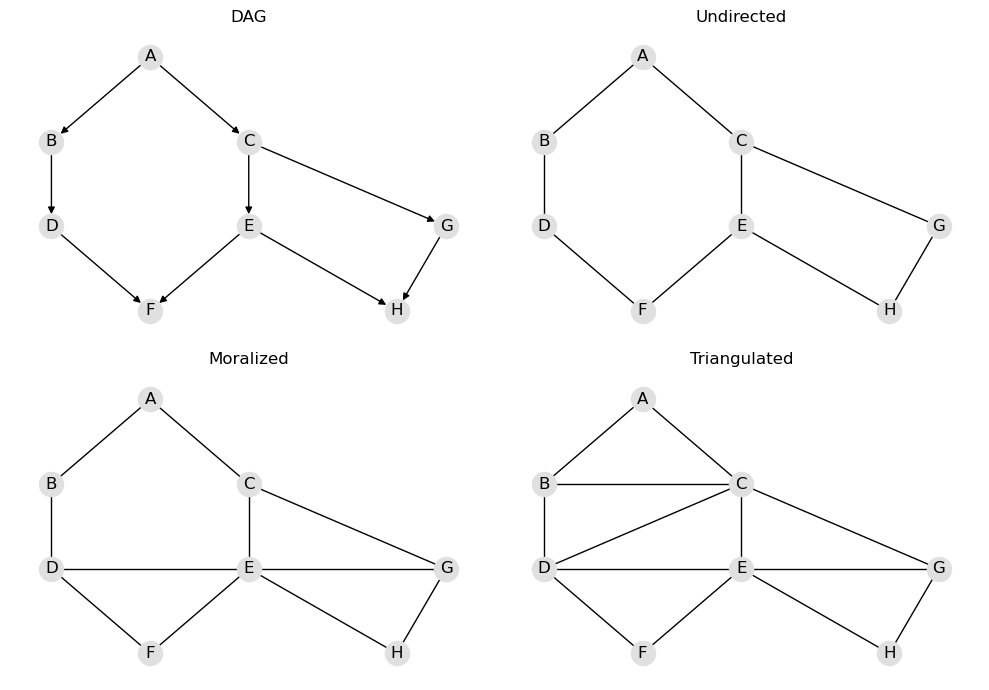
2.2.1. Intermediate graphs
It might be interesting to see the intermediate graphs created. Here’s what the graphs below represent.
DAG: the Huang DAG associated with the Huang BBN
Undirected: the undirected graph of the Huang DAG (directed edges are removed)
Moralized: the moralized graph of the Huang DAG
Triangulated: the triangulated graph of the Huang DAG
[12]:
draw_intermediate_graphs(model)
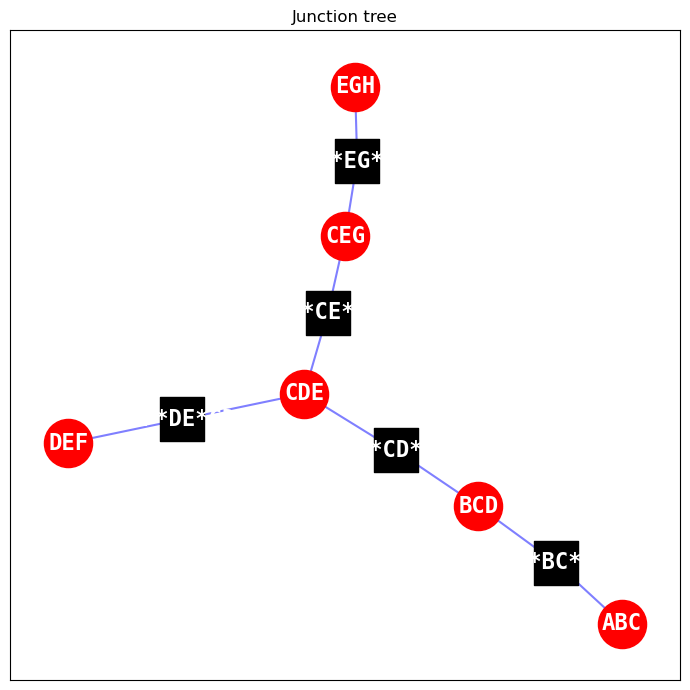
2.2.2. Join tree
The join tree is the main inferencing engine of the reasoning model. We can choose to visualize the join tree too.
red nodes: these are the cliques/clusters of the join tree
black nodes: these are the separation sets of the join tree
[13]:
draw_junction_tree(model)
2.3. Queries
Queries can be classified as follows.
Marginal query (univariate probabilities)
Associational query (conditional probabilities)
Interventional query (causal effect probabilities)
Counterfactual query (counterfactual probabilities)
The above categories of queries follow Pearl’s Causal Hierarchy (PCH) or the Causal Ladder [GDH22].
2.3.1. Marginal query
Let’s just follow one variable, \(H\). The marginal probability of \(H\), \(P(H)\), is easy to query.
[14]:
q = model.pquery()
q["H"]
[14]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.1769 |
| 1 | on | 0.8231 |
2.3.2. Associational query
In an associational query, we want to compute the conditional probabilities
\(P(H|E=\mathrm{on})\), and
\(P(H|E=\mathrm{off})\).
[15]:
q = model.pquery(evidences=model.e({"E": "on"}))
q["H"]
[15]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.322903 |
| 1 | on | 0.677097 |
[16]:
q = model.pquery(evidences=model.e({"E": "off"}))
q["H"]
[16]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.05 |
| 1 | on | 0.95 |
2.3.3. Interventional query
The interventional query is estimating the causal effect one one variable on another. In this case, we want to estimate the causal effect of \(E\) on \(H\). However, \(E\) and \(H\) are confounded through \(\{C, G\}\). We need to apply the do-operator on \(E\) to estimate the causal effect of \(E\) on \(H\). (The causal effect of \(E\) on \(H\) is not simply the associational query we performed above!)
\(P(H=h|\mathrm{do}(E=e))\)
We do not have to control for both counfounders \(\{C, G\}\), it is sufficient to just control for \(C\) to block backdoor paths from \(E\) to \(H\). Thus, using the backdoor adjustment formula, we can compute the causal effect as follows.
\(P(H=h|\mathrm{do}(E=e)) = \sum_C P(H=h|E=e, C)P(C)\)
Let’s compute two causal effects.
\(P(H=\mathrm{on}|\mathrm{do}(E=\mathrm{on})) = \sum_C P(H=\mathrm{on}|E=\mathrm{on}, C)P(C)\)
\(P(H=\mathrm{on}|\mathrm{do}(E=\mathrm{off})) = \sum_C P(H=\mathrm{on}|E=\mathrm{off}, C)P(C)\)
[17]:
p_on = model.iquery(Y=["H"], y=["on"], X=["E"], x=["on"])
p_on
[17]:
H 0.5765
dtype: float64
[18]:
p_off = model.iquery(Y=["H"], y=["on"], X=["E"], x=["off"])
p_off
[18]:
H 0.95
dtype: float64
The average causal effect (ACE) is defined as follows.
\(\mathrm{ACE} = P(H=\mathrm{on}|\mathrm{do}(E=\mathrm{on})) - P(H=\mathrm{on}|\mathrm{do}(E=\mathrm{off}))\)
[19]:
p_on["H"] - p_off["H"]
[19]:
-0.37349999999999983
2.3.4. Counterfactual query
In this counterfactual query, we have the following.
query: \(H\)
factual: \(E=\mathrm{on}, G=\mathrm{on}, H=\mathrm{on}\)
counterfactual: \(E=\mathrm{off}\)
The query is against \(H\); it is the variable we are interested in. The factual is what actually happened. The counterfactual is the hypothetical. Let’s try to word this as a counterfactual statement (not so easy to do).
Given that \(E\) was on, \(G\) was on, \(H\) was on, what is the probability of \(H\) had we set \(E\) to off?
[20]:
Y = "H"
e = {"E": "on", "G": "on", "H": "on"}
h = {"E": "off"}
model.cquery(Y, e, h)
[20]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.043749 |
| 1 | on | 0.956251 |
Let’s try another counterfactual query. We have the following.
query: \(H\)
factual: \(E=\mathrm{off}, G=\mathrm{off}, H=\mathrm{off}\)
counterfactual: \(E=\mathrm{on}\)
As a counterfactual statement, we might naturally say the following (?).
Given that \(E\) was off, \(G\) was off, \(H\) was off, what is the probability of \(H\) had we set \(E\) to on?
[21]:
Y = "H"
e = {"E": "off", "G": "off", "H": "off"}
h = {"E": "on"}
model.cquery(Y, e, h)
[21]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.439756 |
| 1 | on | 0.560244 |
Here’s the last counterfactual query. We have the following.
query: \(H\)
factual: \(E=\mathrm{on}, G=\mathrm{on}, H=\mathrm{on}\)
counterfactual: \(E=\mathrm{off}, G=\mathrm{off}\)
As a counterfactual statement, we might naturally say the following (?).
Given that \(E\) was on, \(G\) was on, \(H\) was on, what is the probability of \(H\) had we set \(E\) and \(G\) to off?
[22]:
Y = "H"
e = {"E": "on", "G": "on", "H": "on"}
h = {"E": "off", "G": "off"}
model.cquery(Y, e, h)
[22]:
| H | __p__ | |
|---|---|---|
| 0 | off | 0.04526 |
| 1 | on | 0.95474 |
2.3.5. Graphical query
Graphical queries revolve around the DAG. Typical graphical queries are about d-separation and discovering counfounders and mediators.
From the DAG, we ask if \(F\) and \(H\) are d-separated, \(I(F, H)\). The answer is false, since there are backdoor paths between \(F\) and \(H\). Here are the backdoor paths between \(F\) and \(H\).
\(F, D, B, A, C, E, H\)
\(F, D, B, A, C, G, H\)
\(F, E, C, G, H\)
\(F, E, H\)
[23]:
model.is_d_separated("F", "H")
[23]:
False
Perhaps we can block all backdoor paths between \(F\) and \(H\) with \(E\), \(I(F,H|E)\)? The answer is false.
[24]:
model.is_d_separated("F", "H", {"E"})
[24]:
False
Can we block all backdoor paths between \(F\) and \(H\) with \(E\) and \(C\), \(I(F,H|E, C)\)? The answer is true.
[25]:
model.is_d_separated("F", "H", {"E", "C"})
[25]:
True
We can query the graph to get us the “minimal confounders” that will block all backdoor paths. There are multiple minimal sets, but with the particular approach we use, the answer is \(\{D, E\}\). The answer to \(I(F,H|D,E)\) is true.
[26]:
model.get_minimal_confounders("F", "H")
[26]:
['D', 'E']
We can use the model itself to 1) get the minimal confounders and then 2) test for d-separation as follows.
[27]:
model.is_d_separated("F", "H", model.get_minimal_confounders("F", "H"))
[27]:
True
If we want a list of all counfounders that can block all backdoors between \(F\) and \(H\), then we can also get that from the DAG.
[28]:
model.get_all_confounders("F", "H")
[28]:
{'A', 'B', 'C', 'D', 'E', 'G'}
2.4. Save
Finally, we can save the reasoning model that we built from the Huang BBN. Note that we are persisting the reasoning model and not the BBN that we depersisted to begin with.
[29]:
from pybbn.serde import model_to_dict
with open("../../_support/bbn/huang-reasoning.json", "w") as fp:
json.dump(model_to_dict(model), fp, indent=1)